From systems operators to systems architects
Going up a level from data generation to think about the data systems we design and embed
We’re moving into an age in which agents are our partners across all aspects of science. Machines will systematically process data, spot patterns, propose experiments, and even generate hypotheses across more and more of our work. I’m excited for how this will accelerate science.
A technological shift of this magnitude requires a similarly big shift in what we study and how we go about it. For one, there should be significantly more emphasis on what data is important to generate and how we build, share, and scale datasets. For another, there should be much more emphasis on funding system architecture that best enables systematic AI-driven discovery, as opposed to primarily funding individual labs to expand datasets.
Today, I’m happy to announce a new $5M funding initiative in structural biology that will experiment with how we make this shift.
The role of scientists in science
Humans remain essential in science. This is a critical moment to stake out where we can most uniquely contribute. What can’t AI solve yet? And for the things it can, how can we leverage its capabilities to do even more creative, generative exploration beyond that radius?
Machines excel at synthesizing large amounts of information through systematic data processing, pattern recognition, and probability calculations. We should replace ourselves in those types of analyses where possible. It’s an uncomfortable, but necessary transition.
There’s still plenty of upstream and downstream work that only we can do:
Downstream: ML predictions are hypotheses, not conclusions. We must test them through research and reuse, closing the loop to validate and improve predictions.
Upstream: Machines work with the data we give them; the nature and design of this data sets the ceiling for what’s possible to predict. We decide what questions to ask and how to architect the systems to answer them.
This piece focuses on the upstream. Too many research proposals focus on generating more data without asking how those expensive exercises will get us somewhere worthwhile. Many brute-force scaling efforts will hit diminishing returns unless we first rethink the kinds of data and data systems we need. This is where human ingenuity will matter most.
The next PDB is the PDB
We need to work smarter, not just harder. As a structural biologist, I’ll give an example close to my heart. And one I’m funding next.
A common trope in funding circles is “What’s the next Protein Data Bank (PDB)?” The question is asked as if structural biology were “done” post-AlphaFold. However, one next big challenge is the PDB; it’s in moving from static protein structures to predicting how they move.
Since form informs function, we use protein structures as clues for what proteins might do in cells. But proteins inherently work through motion: traveling, binding, catalyzing, breathing, and changing conformations. Unlocking protein dynamics would be a giant leap toward uncovering more protein function. This helps us better understand, engineer, and manipulate biology. Biotech companies are already attempting to leverage local data or simulations about protein dynamics for drug modeling.
So one of the “next PDBs” is the PDB itself. It’s not just a remake; it’s PDB 2.0, wherein we can transition from static snapshots to movies. PDB 2.0 can bring protein structures to life.
To get there, we need to think carefully about what conditions are required for such a breakthrough. PDB 1.0’s success depended on being large and standardized, with open data norms. But it also hinged on two other major elements:
Getting the right slice of information
The PDB didn’t contain all protein structures. But it had enough variation in sequences and folds to reveal key design principles. Data was structured for computation (e.g., multiple sequence alignments) to enable breakthroughs. Today, we know more about ML needs, so we can be intentional about focusing our efforts on the data that is most information-rich.
Scaling through embedded practice
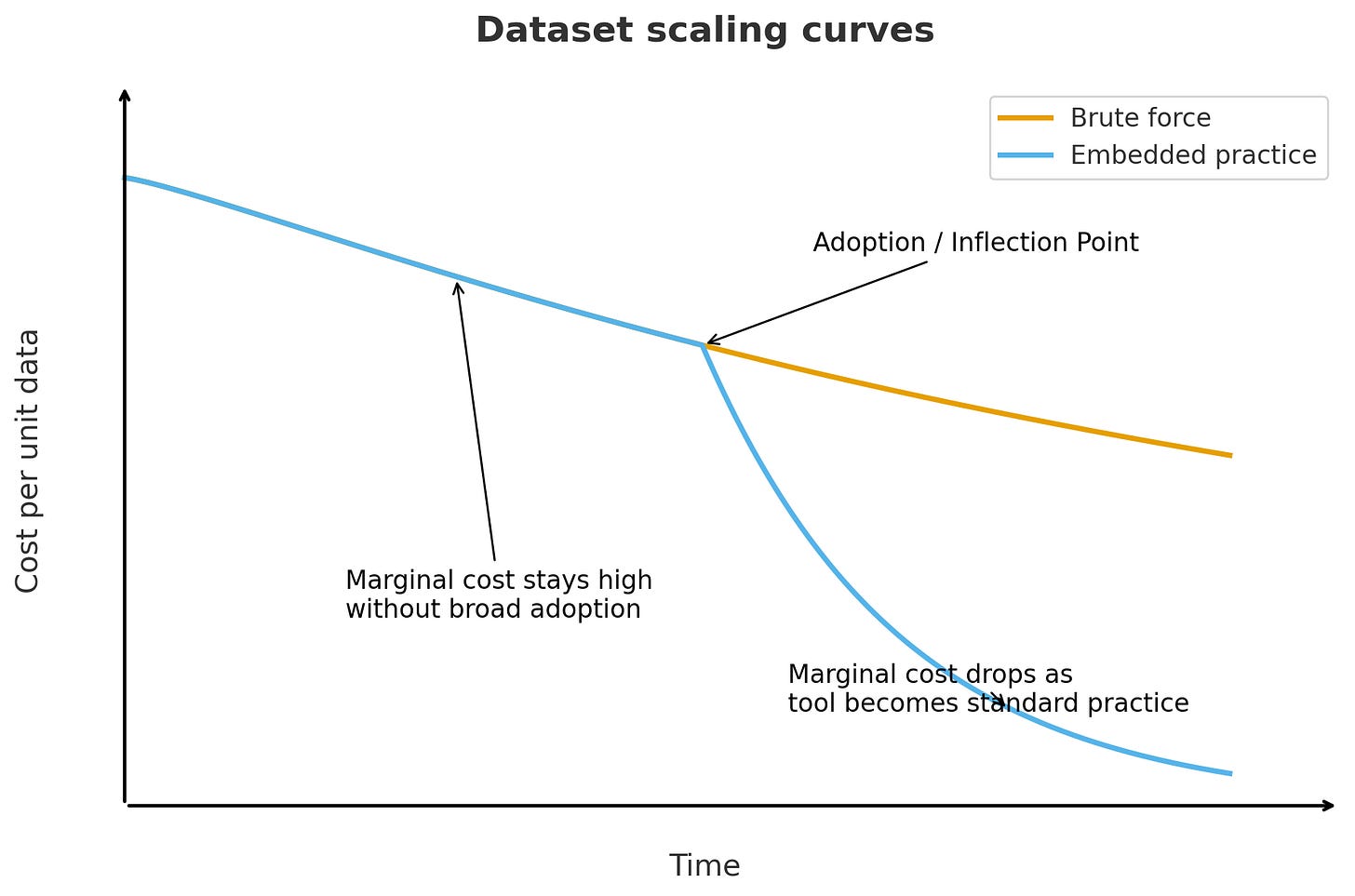
The PDB scaled because crystallography naturally produced standardized data as a byproduct of routine work across methods, hardware, file formats, and infrastructure. This reduced marginal costs as the practice spread. It’s very different from a brute-force approach to data generation, where costs keep rising and the volume of data increases.
(h/t ChatGPT for help with this schematic!)
Finding the right information and embedding it in practice go hand in hand: the right “plumbing” motivates and enables the scientific community to capture valuable data.
We should challenge ourselves to be more cost-effective with PDB 2.0, now that we know more about what we are working towards. We also have machines to help us run quick calculations on what data is most information-dense and therefore most valuable. Instead of taking fifty years and costing tens of billions of dollars, could PDB 2.0 be faster and more efficient?
Principles in practice: introducing The Diffuse Project
With a $5M seed for The Diffuse Project from Astera, work will be coordinated across several universities (UCSF, Vanderbilt, Cornell), scientists associated with national labs and light sources (Los Alamos, Lawrence Berkeley, CHESS), and a team within Astera Institute. Our goal: redesign multiple components of the X-ray crystallography process to capture oft-discarded data on protein conformations.
Traditional crystallography focuses on Bragg scattering, the bright spots that represent the averaged conformation in a protein crystal. Other protein conformations in a crystal produce a more distributed signal — diffuse scattering — which is usually ignored due to its inherent complexity. But today, we have more tools to deal with the messy heterogeneity of diffuse scattering than ever before. Our technical aim is to transform diffuse scattering into a usable signal for hierarchical models of conformational ensembles. I’m excited by how current technologies allow us to better embrace biological complexity in this way.
In close collaboration with experts across the crystallography workflow, we’ve devised a strategy to test whether diffuse scattering could become mainstream practice in structural biology. Our parallel mission is to make the workflow useful, easy, and affordable for anyone studying protein function.
Our team is committed to radical open science by releasing data quickly and sharing progress entirely outside traditional journals — and there have already been many benefits. It has made our brainstorming and collaboration processes much more ambitious and fun. It’s freed us up to focus on outputs that are most useful and representative of our process.
An exciting aspect of the team’s open approach is the chance to experiment in real-time with what scientific publishing could look like in the future. Our open science team is collaborating with our researchers to figure out how they quickly share outputs that are most useful and most representative of our process. As AI becomes more central, our publishing will become more data-centric as well. This is an invaluable opportunity to iterate on this alongside active science to solve real problems on the ground.
We’ve only been in operation for about a month, and we already have data and results to share! You can follow all of this work on The Diffuse Project website.
Utility is the north star
Without journal constraints, we can think more clearly about what will make our science as impactful, accelerated, and rigorous as possible. We keep coming back to the mantra: utility is the north star.
What might high-utility success look like? Our answers span different time horizons, but we strive to be concrete. Here are some examples (which will likely evolve as we go):
Adoption of diffuse scattering by experts outside the initiative
Adoption of diffuse scattering by non-experts
Integration of other modalities with a growing X-ray diffuse scattering dataset
ML models for protein motion trained on diffuse scattering data
Biotech start-ups founded from these models
New therapeutics on the market developed from these models
The diffuse scattering work could take up 7–10 years to reach a scaling inflection point. That’s longer than most projects, but shorter than the time required for the original PDB to scale to a size that enabled AlphaFold. I’m optimistic that we could speed this up if The Diffuse Project truly succeeds in enabling the broader community. It’s a challenge that our team is excited to take on.
A note for funders
Funders, this is the time to flip the script on what science we support. We must move from primarily funding individuals to funding coordinated efforts that “lift all boats” in agent-led discovery systems.
Effective data systems will be a bottleneck for transformational ML. Systems have interdependent parts; if you don’t redesign multiple components at once, you risk getting stuck at a local maximum, due to dependencies between components. This is a well known principle in evolution and engineering. But we’re not great at applying this scientific framework to our own science. We need funding mechanisms that enable sustained, coordinated design sprints across interconnected entities with different areas of expertise.
This kind of systems-level innovation is risky for individual researchers. Not only does it require grappling with the unknown, it also requires grappling with many unknown pieces at once. Any proposal for a project like this would be disingenuous if it provided a super specific roadmap with a high degree of certainty.
Instead, funders need to redefine what success means so that researchers can embrace risk. Namely, teams should prioritize learning and sharing, even about their failures, which can often be informative. The goal can’t be to “win.” It needs to be to learn. The learning process is dynamic (just like proteins!) and we should fund and publish research with that in mind.
Iterating on data systems is also operationally risky. It requires proper infrastructure, engineering, compute access, and ML collaborations. We’re experimenting with unique ways to get this done without creating more bureaucracy or coordination overhead. Astera is providing compute access through Voltage Park and hiring a team for much of the computational and modeling work. This is another avenue by which funders can create an outsized impact through their support.
And at a minimum, we must insist on open science and open data practices. Otherwise, we risk limiting reuse, rigor, and impact. It’s the greatest insurance funders have for a return on their public good investment. Based on my own experiences, I’m very optimistic that we can hold the line on this without sacrificing talent. With the open science requirement, I’ve been able to quickly filter through to some of the smartest scientists, with the most abundant mindsets, with whom to go after big problems. The Diffuse Project’s team is awesome. I could not be more thrilled about the stellar set of scientists leading the research.
A first step towards the future
The Diffuse Project is not an outlier. It’s one example of the broader shift we need to make towards a data-centric future of discovery.
This shift will allow us scientists to work smarter. As machine learning systems become more prominent in hypothesis generation, simulation, and interpretation, the role of scientists will not disappear — it will evolve. Our ability to be creative about what data we generate, how we go about it, and what it enables, will become an increasingly central part of how we contribute to discovery.
This is not a loss. It’s an opportunity. It’s a chance — for both scientists and funders — to reclaim the architectural layer of science as a thoughtful, creative, and essential realm of experimentation and discovery. And it’s a place where human judgment still matters — perhaps more than ever.
If you’re working on something that could benefit from this kind of innovation, I’d love to see it published openly so I can follow and learn through open discourse. In the spirit of open science, I generally prefer not to respond to private emails or DMs. I would love to see open science publishing in effect at the ideation stage too.

I'm not a specialist in structural biology so I apologize if my question is naive. But I had questions about the proposed technique.
If I understand your methodology, you take the images taken in X-Ray crystallography that don't conform to the majority of other images, the idea being that you may find different conformations that indicate other valid confirmations which can reveal how the protein moves. My question is: how do you differentiate images of valid conformations from proteins that broke apart in the purification step, denatured proteins, etc? And how do you validate the output of your process?
“exercises that will get us somewhere worthwhile”
Well said!